Vizgen Advances Spatial Genomics Research with New MERFISH Data Release Program
Vizgen releases new dataset generated by Vizgen’s MERSCOPE™ which includes a high-resolution spatial map of GPCR and RTK expression within the mouse brain
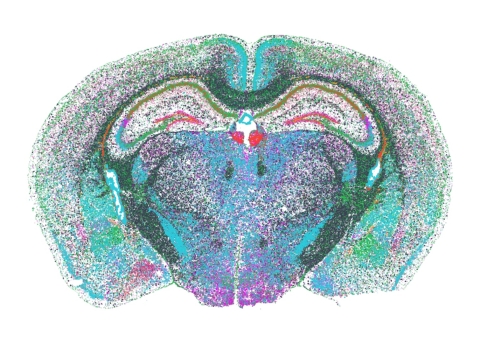
The Vizgen MERFISH Mouse Brain Receptor Map dataset contains 554,802,908 transcripts and 734,696 cells representing the largest dataset of single-cell spatial transcriptomics publicly available to date. (Photo: Business Wire)
May 06, 2021 06:00 AM Eastern Daylight TimeCAMBRIDGE, Mass.--(BUSINESS WIRE)--Vizgen, the life science company visualizing biology to advance human health through the first complete solution for single-cell spatial genomics, today released its first freely available dataset, the Vizgen MERFISH Mouse Brain Receptor Map. This is the first of a series of datasets that will be made available through the company’s new Data Release Program.
“Our goal is to demonstrate the unique power of MERSCOPE’s data while providing a valuable resource for the research community.”
Tweet thisVizgen’s release of the MERFISH Mouse Brain Receptor Map marks the first time the research community at large can access open-source spatial genomics data. This will allow researchers to truly understand the power of spatial technologies, while also experiencing the capabilities and advantages of unparalleled scope and resolution enabled by MERSCOPE. The Vizgen MERFISH Mouse Brain Receptor Map contains the exact position of transcripts from 483 genes across three full coronal slices with three biological replicates for each slice. In all, the whole dataset contains 554,802,908 transcripts and 734,696 cells representing the largest dataset of single-cell spatial transcriptomics publicly available to date. The data set also includes cell boundary polygons, and high resolution DAPI and Poly T mosaic images.
Through the data, researchers can learn how MERFISH technology provides a direct window into the molecular, cellular, and functional organization of intact biological systems and gain greater insight into the mechanisms underlying both health and disease. This and future datasets will be accessible through a form on Vizgen’s website. To eliminate barriers to access, datasets released through the Vizgen MERFISH Data Release Program will be freely available to participants to use as they wish.
Notably, the dataset provides unprecedented detail about the spatial expression of hundreds of receptor genes – notoriously difficult data to capture due to many of these genes being expressed at very low levels within the tissue.
The mission of the Vizgen Data Release Program is to provide researchers early access to MERFISH data generated on the MERSCOPE platform, which includes images and tables that describe the exact spatial position and their colocalization within individual cells. Each dataset released will represent a different “cell atlas,” expanding on the global initiative to generate systematic and comprehensive reference maps of cells and how they function together in both normal and diseased biological tissues. Overall, through its Data Release Program, Vizgen seeks to accelerate biological discovery by making MERFISH data available to scientists of all backgrounds and skillsets.
“We are excited to make this data publicly available so that the research community can immediately leverage the high-quality spatial genomics measurement of MERSCOPE to gain a deeper understanding of the complexity of these biological systems. Each dataset released will have a different anatomical or disease-associated focus and will be a resource for illuminating open biological questions that are difficult to address without a highly multiplexed, spatial genomics technology,” said George Emanuel, Vizgen co-founder and Director of Partnerships and Technology. “Our goal is to demonstrate the unique power of MERSCOPE’s data while providing a valuable resource for the research community.”
About MERFISH
MERFISH is a massively multiplexed single molecule imaging technology capable of simultaneously measuring the copy number and spatial distribution of hundreds to thousands of RNA species across hundreds of thousands of individual cells. The unique approach produces unmatched detection efficiency and images with nanometer-scale resolution.
Vizgen’s MERSCOPE is the only commercially available platform designed for running MERFISH experiments, and the first complete platform for single-cell spatial genomics. The instrument is flexible and is amenable to virtually any sample or tissue type. MERSCOPE is accompanied by reagents and consumables, fully automated analytical instrumentation, and data visualization software. Together, these enable combinatorial labeling, sequential imaging, and error-robust barcoding for the spatial profiling of hundreds of individual transcripts within hundreds of thousands of cells in a single instrument run.
Cells within biological systems typically follow a distinct spatial organization that is fundamental to their functionality. As such, spatial context is essential to classifying individual cell types understanding how they influence one another. Vizgen’s MERFISH Mouse Receptor Map focuses on neural tissue, but this is just one of many research areas that requires spatial context to fully approach yet-unanswered questions.
About the Vizgen MERFISH Mouse Brain Receptor Map
The Vizgen MERFISH Mouse Brain Receptor Map demonstrates the strength of MERFISH to characterize difficult to profile, lowly expressed but functionally relevant genes across large tissue areas.
One highlight of this dataset is that it offers spatial information about nonsensory G-Protein coupled receptors (GPCRs): key mediators of signaling between neurons. In contrast to sensory GPCRs that directly receive inputs from the external environment, around 90 percent of the ~370 nonsensory GPCRs expressed in the brain are critical for the regulation of neuronal excitability, metabolism, reproduction, development, hormonal homeostasis, and behavior. Furthermore, studies are finding GPCR signaling to play an increasingly prominent role in processes underlying aging of the brain, particularly in how GPCR signaling over long periods of time effects the central nervous system and the progression of neurodegenerative disorders. 1 As such, 30 percent of marketed drugs modulate specific GPCRs. 1, 2, 6 Yet, analysis of nonsensory GPCRs in vivo is a difficult process because of their structural properties, low abundance, and lack of highly specific antibodies. 2 Some GPCRs to highlight in the Mouse Brain Receptor Map that are functionally relevant but lowly expressed include Oxtr (oxytocin receptor )3, Tshr (Thyroid stimulating hormone receptor)4, and Insr (insulin receptor).5
MERFISH technology can now play a critical role in the continued mapping of GPCR expressions within the brain by providing high throughput, spatially resolved information with robust sensitivity and resolution to gain greater insight into the structure of the healthy brain and the phenomenon of neurological decline.6
Specifically, the Vizgen MERFISH Mouse Receptor Map contains:
- Gene panel: 483 total genes including canonical brain cell type markers, GPCRs, and receptor tyrosine kinases.
- Samples: Measurements of three full coronal slices across three positions in the brain with three biological replicate measurements for each position.
- Output data for each measurement: a list of all detected transcripts, gene counts per cell matrix, additional spatial cell metadata, cell boundary polygons, and high resolution DAPI and Poly T mosaic images.
This dataset offers the same level of detail as recent data 7 coming out of the lab where MERFISH was developed, led by Dr. Xiaowei Zhuang, a Howard Hughes Medical Institute Investigator, and David B. Arnold, Jr. Professor of Science at Harvard University. Jiang He, PhD, Director of Scientific Affairs stated, “Vizgen is providing a robust solution for analyzing hundreds of thousands of cells simultaneously that enables identification of rare cell types and lowly expressed genes. The unprecedented view of the spatial organization that MERSCOPE provides at the subcellular level will allow researchers to gain transformative insights unraveling the complexity of biological systems.”
About Vizgen
Vizgen is developing the next generation of genomic profiling tools that enable researchers to gain new insight into the biological systems that underlie human health and disease with spatial context. The company's MERSCOPE platform enables massively multiplexed, genome-scale nucleic acid imaging with high accuracy and unrivaled detection efficiency at subcellular resolution. MERSCOPE provides transformative insight into a wide range of tissue-scale basic research and translational medicine in oncology, immunology, neuroscience, infectious disease, developmental biology, cell and gene therapy, and is an essential tool for accelerating drug discovery and development. For more information, go to www.vizgen.com and connect on social media @Twitter, @LinkedIn and Facebook.
References
- G-Protein-Coupled Receptors in CNS: A Potential Therapeutic Target for Intervention in Neurodegenerative Disorders and Associated Cognitive Deficits (2020) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7072884/
- In vivo mapping of a GPCR interactome using knockin mice (2020) https://www.pnas.org/content/117/23/13105
- Oxytocin pathway gene networks in the human brain (2019) https://www.nature.com/articles/s41467-019-08503-8
- The expression of thyrotropin receptor in the brain (2001) https://pubmed.ncbi.nlm.nih.gov/11159854/
- Igf1R/InsR function is required for axon extension and corpus callosum formation (2019) https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0219362
- Expression map of 78 brain-expressed mouse orphan GPCRs provides a translational resource for neuropsychiatric research (2018) https://www.nature.com/articles/s42003-018-0106-7
- Molecular, spatial and projection diversity of neurons in primary motor cortex revealed by in situ single-cell transcriptomics. (2020) https://www.biorxiv.org/content/10.1101/2020.06.04.105700v1
Contacts
Brittany Auclair
Vizgen
Brittany.auclair@vizgen.com
Editor Details
-
Company:
- CG Life
-
Name:
- Kathleen Proudfoot
- Email:
-
Telephone:
- +17087176570
- Website:
Related Links
- Website: Press Release